Spatial transcriptomics with LCM-Seq to study fluorescently tagged neurons from transgenic mice
LCM-Seq protocol developed for whole-transcriptome profiling of spatially resolved cholinergic neuron populations
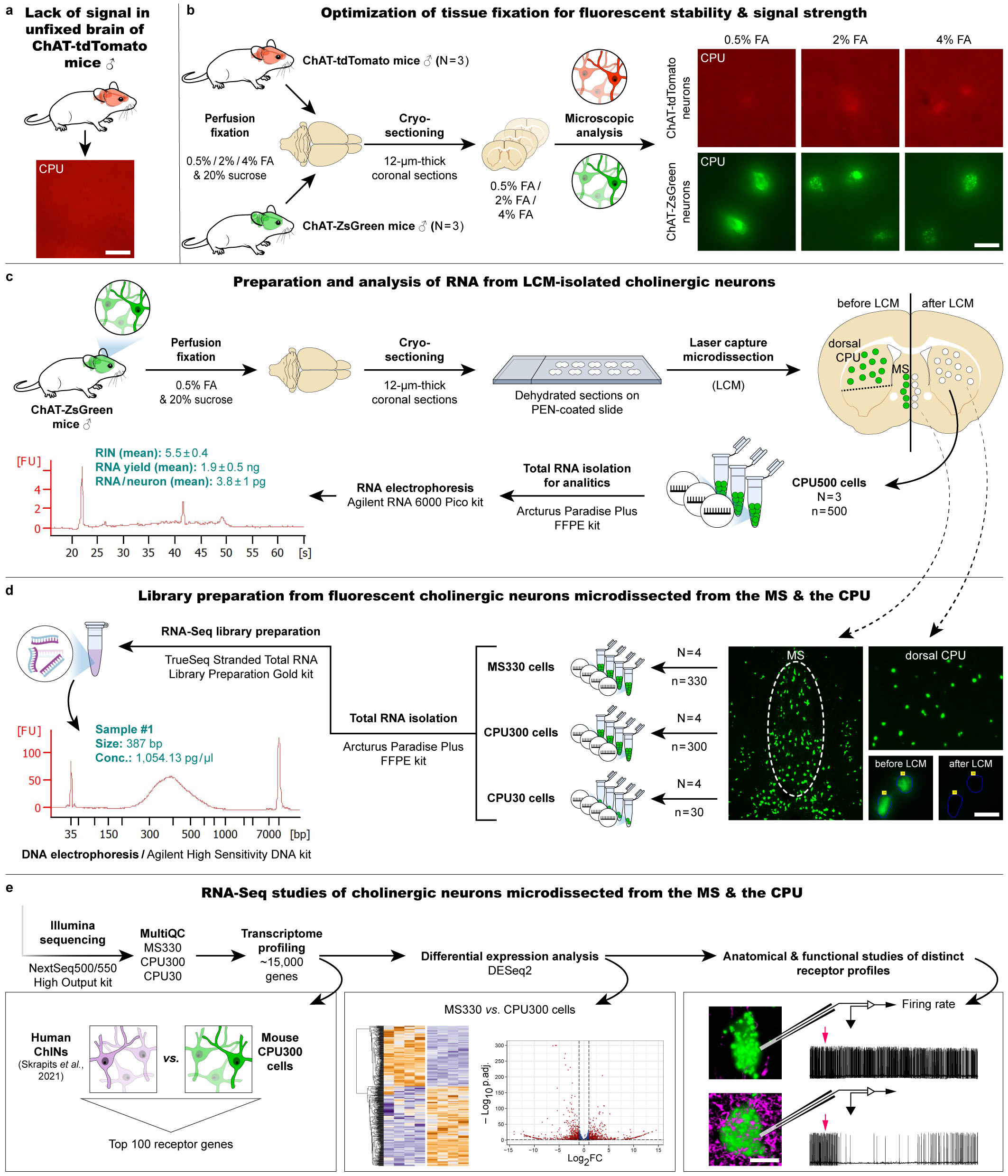 This cutting-edge method developed in our laboratory represents a widely used approach in nearly all ongoing research projects of our team. Selection of the fluorescent marker protein and optimization of the fixation, RNA isolation, RNA-Seq library preparation protocols allows us to detect and quantify with high precision ~ 15,000 different transcripts from neuronal pools collected with laser capture microdissection (LCM). Bioinformatic analysis provides deep insight into the gene expression profiles of cholinergic neurons from the caudate putamen (CPU) and medium septum (MS) regions, revealing also molecular differences between these two.
This cutting-edge method developed in our laboratory represents a widely used approach in nearly all ongoing research projects of our team. Selection of the fluorescent marker protein and optimization of the fixation, RNA isolation, RNA-Seq library preparation protocols allows us to detect and quantify with high precision ~ 15,000 different transcripts from neuronal pools collected with laser capture microdissection (LCM). Bioinformatic analysis provides deep insight into the gene expression profiles of cholinergic neurons from the caudate putamen (CPU) and medium septum (MS) regions, revealing also molecular differences between these two.
Rumpler, Göcz et al., J Biol Chem. 2023 Sep;299(9):105121
This approach has been used successfully in studies of the hypothalamic neuronal network regulating reproduction.
Göcz és mtsai. Proc Natl Acad Sci U S A. 2022 Jul 5;119(27):e2113749119
Göcz és mtsai. Front Endocrinol (Lausanne). 2022 Aug 25;13:960769
With adaptations, a similar protocol allowed us to carry out RNA-Seq studies of cholinergic interneurons (identified by cell size) from the human putamen.
Skrapits és mtsai., Elife. 2021 Jun 15;10:e67714






