Why is it good to know the transcriptome? The importance of a method and the benefits of the results
The Reproductive Neurobiology research group led by Erik Hrabovszky has published a methodological breakthrough in the Journal of Biological Chemistry. We spoke to Eva Rumpler, who, along with Balázs Göcz, is the shared first author of the paper.
The choice of methods used in experiments is a crucial element of any work. "Die Methode ist alles" ("The method is everything"), said Karl Ludwig, a renowned 19th century German physiologist, who was elected a member of the Hungarian Academy of Sciences in 1872, but a hundred years later our world-famous neuroscientist János Szentágothai, former president of the Academy, also emphasised the importance of the methods used in research. He was convinced that reliable scientific results can only be achieved through solidly based methods, and that innovative techniques are an indispensable tool for knowledge in fields such as neurobiology. It is likely that he would have welcomed the development of methods to identify and determine the complete RNA pool of nerve cells of any species by the research team led by Erik Hrabovszky.
- What is a transcriptome and why did you study neurons containing gonadotropin-releasing hormone (GnRH) in the brain?
Évi Rumpler
- All the cells that make up the body contain the same set of DNA, the difference between cell types lies in the diversity of RNA molecules that are transcribed from DNA. The RNA transcript that is specific to each cell type is called a transcriptome. This RNA transcript, in addition to being dependent on the type of cell and its anatomical location, is constantly changing throughout our lives.
The second part of the question is actually answered by the name of our group - Reproductive Neurobiology - because the neurons in the brain that contain gonadotropin-releasing hormone (GnRH) regulate reproductive functions. The GnRH peptide binds to the GnRH receptors of the pituitary gland, causing the
release of the hormones LH and FSH, which regulate gametogenesis and the
production of sex steroid hormones in the reproductive organs.
- Since when have you been trying to understand the RNA repertoire, or gene expression profile, characteristic to GnRH neurons?
- Quite a long time, as have many others besides us. Early methods were characterized by low throughput and low sensitivity. Later chip-based techniques have revealed much more about the RNA milieu of cells, but far from everything.
- Last year you published a methodological article on this topic, also in JBC!
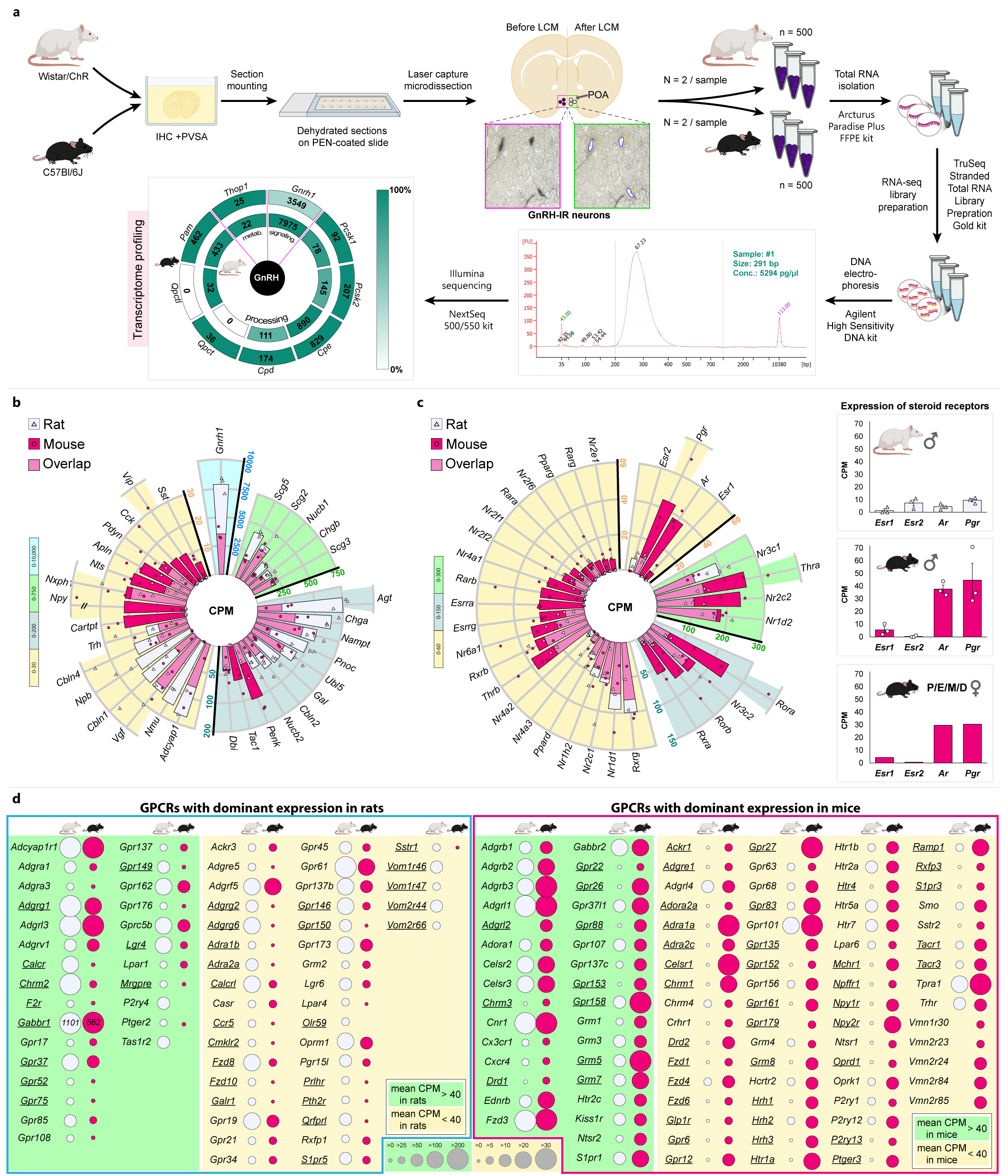
- That article is a close precursor to our current research. We used a new generation sequencing approach to determine the RNA set from neurons extracted from histological sections by laser microdissection (LCM), developed under Erik's leadership. This method, named "LCM-Seq" for the abbreviation of the technique used, worked reliably, but its more general applicability has a drawback. The visualisation of neurons, a prerequisite for the use of laser microdissection in this method, still required the use of genetically engineered mice that produce fluorescent protein in the neuron type to be studied.
- The creation of the genetically modified mouse model itself is extra time, and it is obvious that studying other species is not always possible!
- That is why we started to study the neurons identified by immunohistochemistry in formalin-fixed tissues. With the newly developed method, we have successfully mapped the entire mRNA repertoire of GnRH neurons regulating reproduction in rats, in addition to mice. Perhaps the most exciting aspect is that, with minor modifications, this method is already suitable for mapping the transcriptome of certain human neuron types from post-mortem histological samples.With the first authorship of Szabolcs Takács, our manuscript describing a technique suitable for the study of human neurons is almost ready!
- I myself am looking forward to the results, since, as they say, we do not want to know everything about mice and rats, nor is their cure our primary goal.
In your recently published article, you show a kind of neuronal RNA set in both mice and rats. What can you learn about a species from that?
- We are able to identify and quantify between 14,000 and 16,000 different mRNAs in a single cell type. Thanks to this large number, the method is also quantitative. It is possible to identify genes with variable expression and to detect small shifts in the transcriptomic map in a given experimental setup with high accuracy.
- The fact that an RNA is detectable does not guarantee that protein synthesis has taken place. Are there valid conclusions to be drawn from studying the mRNA pool with respect to protein levels and protein function?
- Obviously, the result requires confirmation, further experiments. I can give one of our recent results as an example. The transcriptome of GnRH neurons expressed high levels of mRNA encoding the neurotensin peptide type 2 receptor. Based on the transcriptome result, we hypothesized that neurotensin significantly affects the function of GnRH neurons. My colleague Imre Farkas supported this hypothesis with electrophysiological studies.
- What else can transcriptomics be used for?
- It can also be used to study regulatory/regulatory RNAs. This is a rapidly developing, relatively new field of science. Such regulator RNAs do not encode proteins but play an important role in regulating cell function.
- What has been the biggest challenge in your method development experiments?
- Preserving the integrity of the RNA molecules during immunohistochemical labelling and storage of the sections, as well as organising the experiments. Fortunately, Balázs Göcz helped me with this. I was working part-time while on GYED, so he continued the experiments I started at the beginning of the week and finished them in the second half of the week.
- How did the mixture that was able to preserve RNA come together and how were the experiments carried out?
- Ideas and concepts for RNA preservation were taken from the almost forgotten old literature and reviewed in the lab. We have had many failed attempts, but these have not been counted in hindsight. One thing is for sure: developing the final protocol was a long process.
- A method published in a scientific journal should work in any laboratory. What advice can you give to those who are preparing to do similar work? What should they pay most attention to?
- It may not be surprising that I emphasize precision and cleanliness. Even in the case of RNA experiments, it is only at the end of a long process that we can be sure that we have done the right job. It is essential to strictly follow the protocol, as increasing the concentration of RNA-preserving additives can negatively affect the detectability of immunohistochemical signals!
- What was the most surprising result?
- One of our observations, which contradicts the literature, was that only Esr1 mRNA encoding the oestrogen receptor α is expressed in mouse GnRH neurons and only Esr2 mRNA encoding the oestrogen receptor ß in rat. So there is a difference between the two species that few expected. The other equally important observation is the important role of the neurotensin receptor in the stimulatory regulation of GnRH neurons, mentioned earlier.
- Your method can be used to determine the transcriptome of cells other than neurons. Are there any obstacles to its widespread use?
- It may be somewhat limited by the instrumentation requirements of laser microdissection (LCM) for cell isolation and the high price of a specialised cDNA library kit. For us, the most important application is the determination of the transcriptome of various cell types collected from post mortem human brain tissue, which could help to identify new targets suitable for treating various diseases.
- What is the subject of your current experiment?
- Among others, we investigate the effects of sex steroid hormones on gene expression. The feedback of estrogen and androgen hormones in the brain plays a crucial role in the central nervous regulation of reproduction. Our goal is to gain a more precise understanding of the molecular mechanisms underlying this phenomenon. We also plan to apply our new method, as it may provide an opportunity to better understand previously unexplored aspects of these regulatory processes.